A next generation microarray technology originally developed for the cannabis sector could help the poultry industry better detect and monitor for the presence of Salmonella and other pathogens.
Salmonella in poultry is under intense scrutiny right now. In the Fall of 2022, the U.S. Department of Agriculture (USDA) Food Safety and Inspection Service (FSIS) unveiled a proposed framework to improve Salmonella control in poultry products. This includes stricter monitoring protocols for specific Salmonella serotypes.
Salmonella is one of the most common causes of foodborne illnesses in people. With over 2,600 known serotypes, however the individual serotype can be difficult to identify.
“Our platform will actually test for 13 of these, including the three that need to be traced,” said Carol Dugas, global director of sales, PathogenDX.
These serotypes include Enteritidis, Typhimurium, Newport, Javiana, Heidelberg, Muenchen, Saintpaul, Montevideo, Infantis, Braenderup, Oranienberg and Thompson
How it works
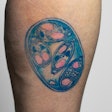
The microarray can identify genes that are highly correlated to certain Salmonella serotypes using multiplexed end point PCR reaction.
“During that reaction, we’re detecting those specific genes. If that gene is detected, we amplify our specific target and then the target is also labeled in our PCR reaction. That labeled amplifier is hybridized on to our microarray,” Shaun Stice, senior scientist, PathogenDX, explained.
In other words, to identify the Salmonella serotype, set up a simple PCR reaction, run it in a thermal cycler and add a buffer to it. Once this is run through the microarray technology, an online portal system interprets and shares the results within 24 hours.
“The traditional microarray people envision is one that’s a little more complex,” Stice added, noting that the overnight hybridization typically takes an hour.
Updates to the microarray technology mean that hybridization is performed in less than an hour at room temperature. In the future, the microarray could potentially monitor for other pathogens, including Campylobacter and avian influenza.